



Interactive tools to search transcripts, browse Gene Ontology, and view KEGG pathways derived from ginger challenged with Pythium myriotylum (P0822).

Three complementary entry points for exploring the dataset.
Key summaries from differential expression and enrichment analyses.
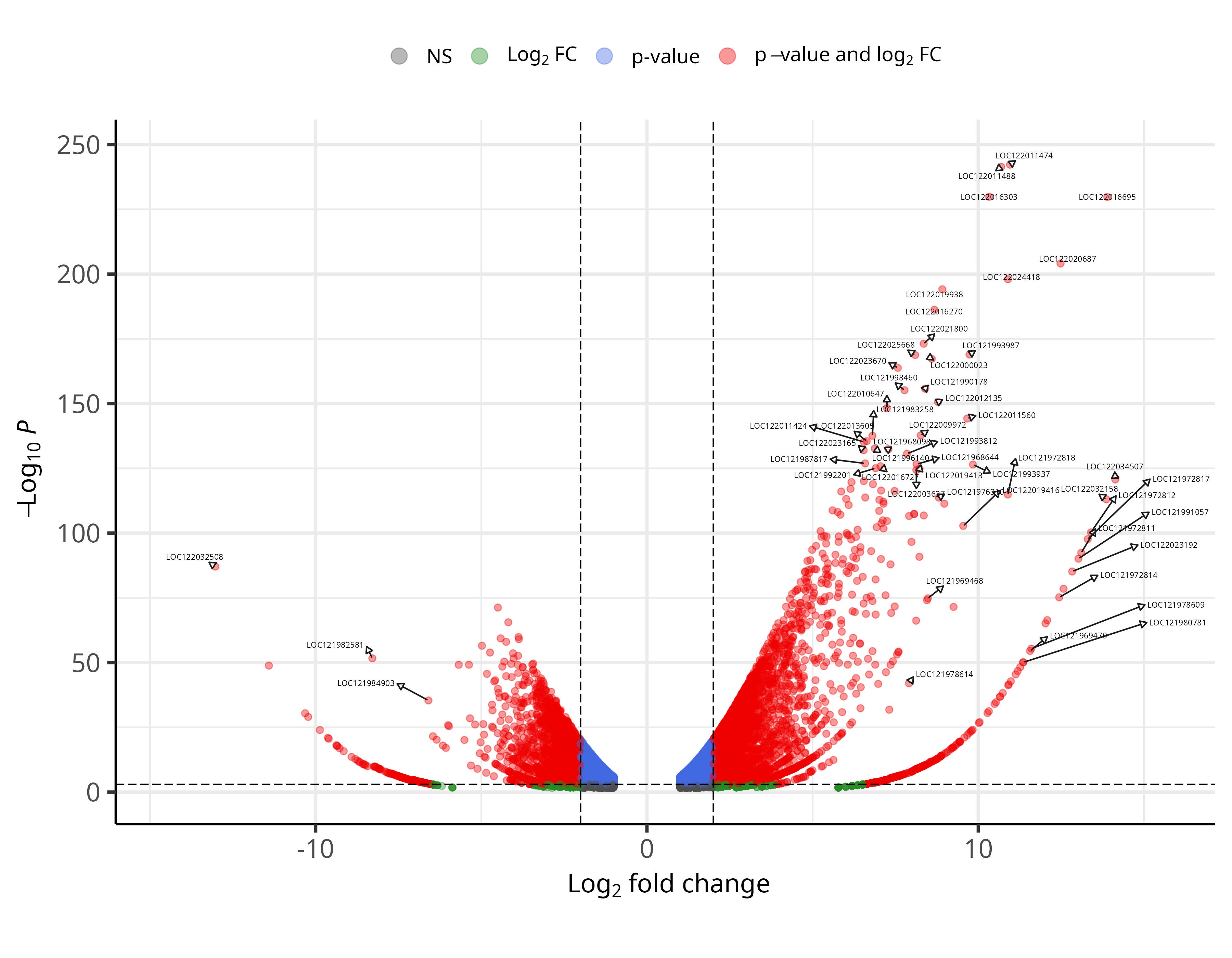
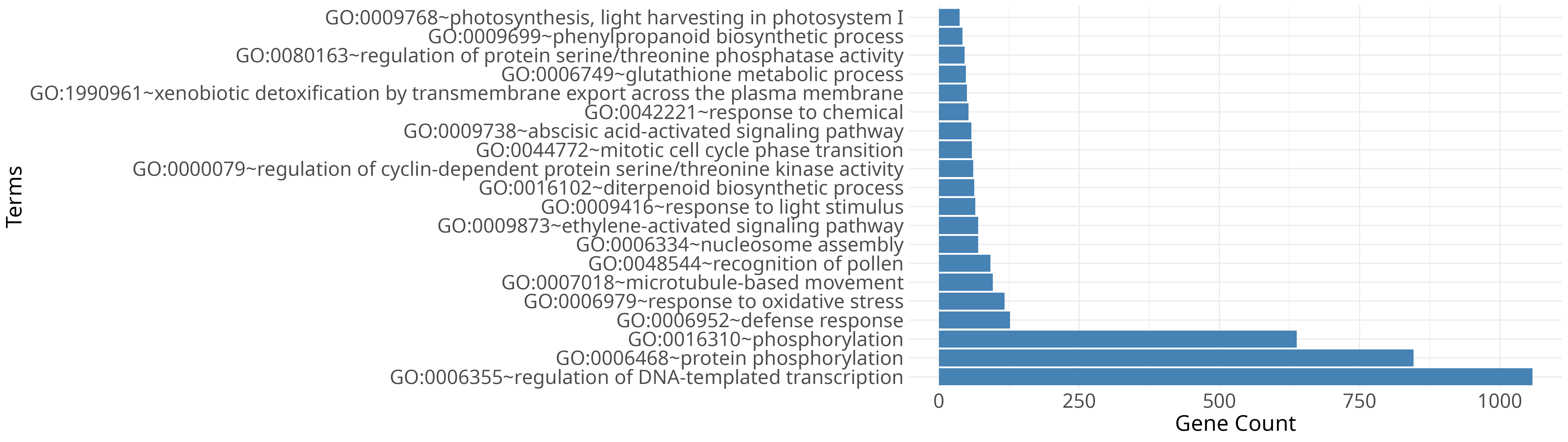
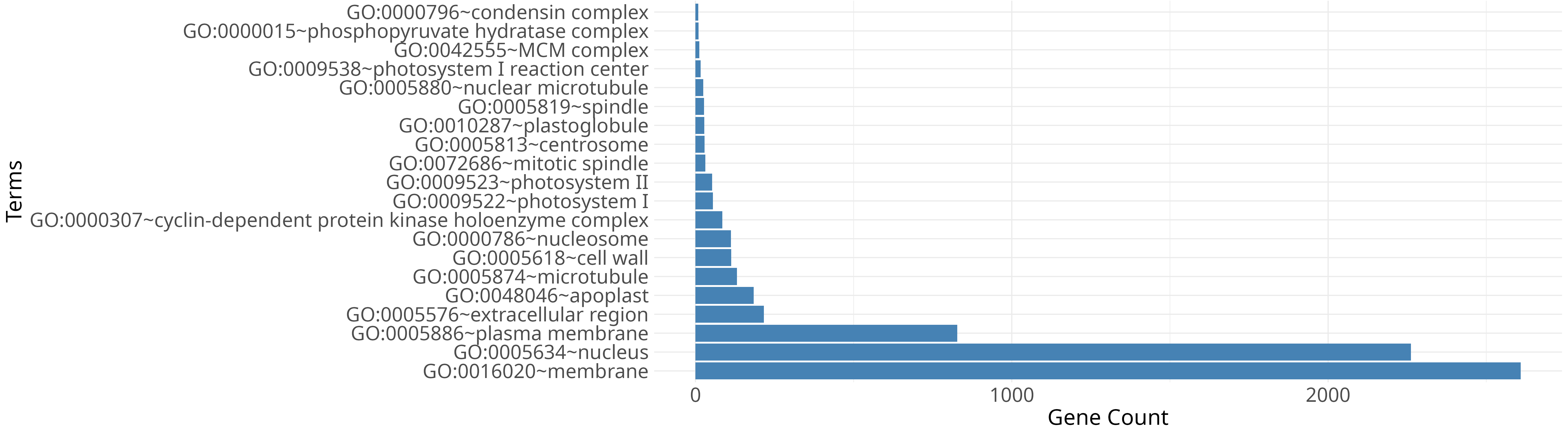

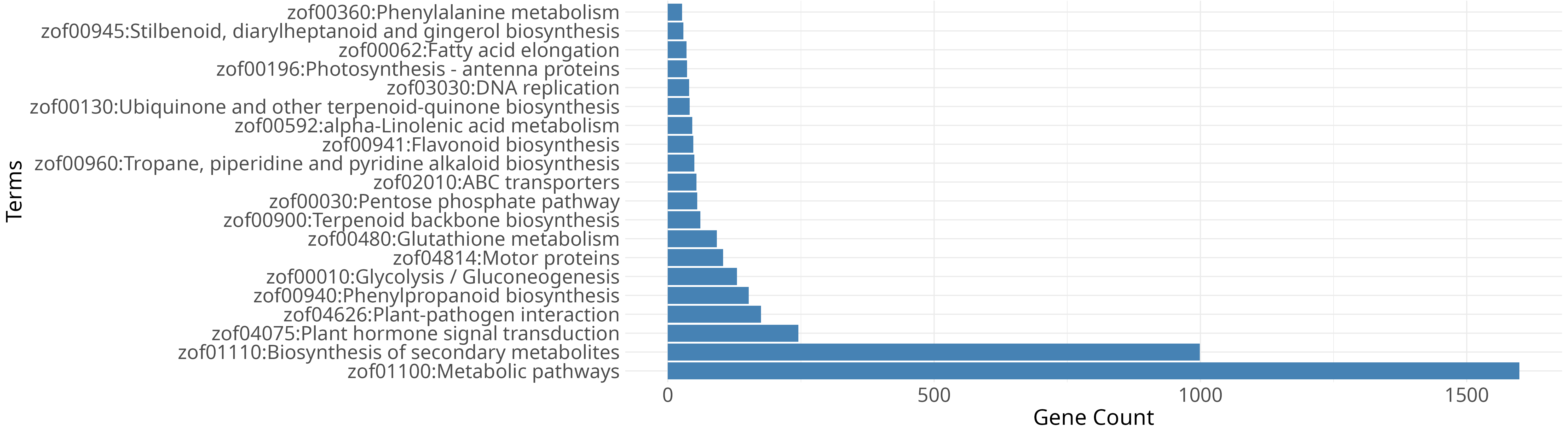
Ginger (Zingiber officinale) is one of the widely cultivated spice crops, yet highly susceptible to soft rot caused by Pythium spp., leading to substantial yield losses worldwide. The GiPyDB consolidates transcriptome data from ginger inoculated with Pythium myriotylum isolate P0822, enabling exploration of infection responsive transcriptional changes.
This database centralizes infection‑responsive transcripts in ginger to support research in plant–pathogen interactions and molecular breeding for disease resistance.
The work was supported by the ICAR project, ICAR-CIB IV: Enabling Climate Resilience and ensuring food and Nutritional security through Genome Editing in Horticultural Crops [Sub project: - Application of genome editing to develop trait-specific varieties in ginger]